Description
In this project, we employed ICA to model the gene expression data for gene selection, and then applied NMF and its extensions, i.e., SNMF and NMFSC to cancer clustering using the selected genes. The proposed method was validated on the leukemia dataset, embryonal tumors dataset from the central nervous system, and the medulloblastoma dataset.
It can be found that improved clustering results were achieved by selecting the key genes using ICA. From the experimental results, we can see that the ICA-based gene selection is useful to detect the subsets of relevant genes for tumor clustering, especially when coupled with the NMF clustering method. It should be noted that although the three datasets used in our experiments have similar number of genes, i.e., about 5000, our method has no constraints on the number of genes contained in the data. In fact, our proposed method can be applied to the datasets that have much more genes.
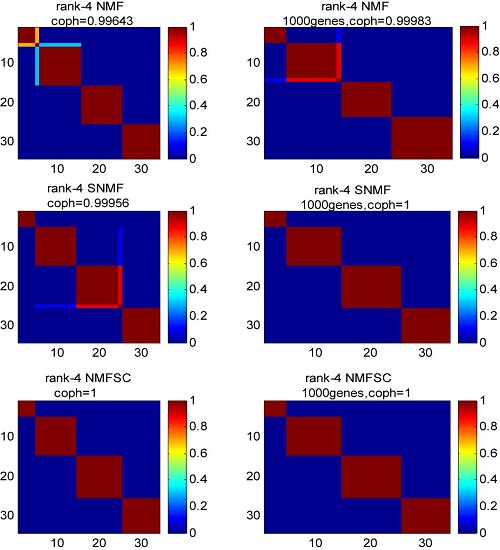
Reordered consensus matrices and the corresponding cophenetic correlation
ref :
Zheng, Chun-Hou, De-Shuang Huang, Lei Zhang, and Xiang-Zhen Kong. “Tumor clustering using nonnegative matrix factorization with gene selection.” IEEE Transactions on Information Technology in Biomedicine 13, no. 4 (2009): 599-607.



Reviews
There are no reviews yet.