Efficient Cell Segmentation and
Tracking of Developing Plant Meristem
Introduction
Proper understanding of the causal relationship between cell growth patterns and gene expression dynamics is one of the major topics of interest in developmental biology. Information such as rates and patterns of cell expansion play a critical role in explaining cell growth and deformation dynamics. The need for quantification of these biological parameters is important to biologists. However manual analysis is extremely tedious because of the high dimensionality and complexity of data. The subject of this study, the shoot apical meristems (SAMs) also referred to as the stemcell niche, is the most important part of the plant body plan because cells for all the above ground plant parts are supplied from it. At the same time, the size of the stem-cell niche remains stable in spite of a continuous displacement and diversion of cells into a differentiation program. Under-standing this dynamics is a major research thrust for developmental biologists. In this work, we focus on studying the developing plant meristem with the goal of obtaining very accurate segmentation and tracking. Confocal laser scanning microscopy is used to observe given set of SAMs labeled with plasma membrane localized yellow fluorescent protein (YFP), repeatedly for about 3 days by taking serial images at every 3 hour intervals. At each time point, multiple images are obtained at different depths. Each stack from all time points is registered by method of maximization of mutual information. As a result, the images taken from different time points but at the same slice are registered. Fig.2 shows an example of the time lapse images. To keep the plant alive for a long period of time, it is necessary to limit its exposure to the laser. This results in poor image quality. This presents significant challenges to image analysis since the segmentation and the tracking needs to be robust to the poor image quality.
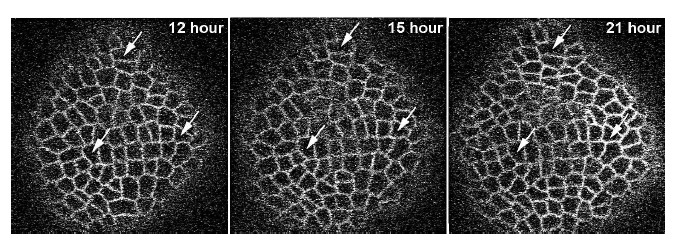
Figure 2: Example of three images at three time instants. Arrows show same set of cells at time points noted on each panel.points noted on each panel.
Segmentation
The authors in used a level set algorithm for segmentation. Since the input raw images have low signal to noise ratio in the periphery and also in the central regions of the plant stem, the segmentation results of this work are of poor quality. We used watershed transformation to segment cell boundaries. Watershed treats the input image as a continuous field of basins (low intensity pixel regions) and barriers (high intensity pixel regions), and outputs the barriers, which are the cell boundaries of all the cells in the image. Prior to applying the watershed algorithm, the input image from the confocal microscope undergoes low pass filtering. This Gaussian filtered image is further processed using H-minima transformation in which all the pixels below a certain threshold percentage h are discarded. These two steps minimize the effects of noise in our segmentation. The threshold value h plays a very crucial role in the watershed algorithm. Generally, a higher value of the threshold parameter h performs under segmentation on the image, and inversely a lower value over-segments it, as shown in Fig. 3.

Figure 3 : The detection of cell boundaries varies with the different values of watershed threshold h. First image is the raw input image. Over-segmentation using watershed is shown in the middle image, and under-segmentation in the right most image.
propriate threshold value such that only the correct cell boundaries are detected by watershed. One of the main contributions of this work is a quantitative metric to evaluate the “correctness” of the segmentation. We observe that the area of all the cells in a plant is almost uniform in the image. Thus watershed should ideally produce a segmented image that contains similar sized cells. We use variance in the area of the cells as a metric to measure correctness of segmentation. Thus, the value of h should be chosen such that variance of area in the segmented image is minimum. Since it is a quantitative metric, the optimal value of h is found automatically without manual intervention. The appropriateness of this metric is also verified through the successful use of these segmented images in tracking with significantly improved results. Figure 4 illustrates the automatic evaluation of h for the given sample image. The segmented image obtained after watershed segmentation is a binary image. Comparison with: Since the tracking algorithm uses cell properties such as area of the cell and its centroid, it is important to retain the shape and structure of cells in the segmented images. In a noisy image, level-set method fails to preserve these cell properties, which leads to incomplete

Figure 4: The graph shows the watershed threshold h in the X axis, and the variance in the area of the segmented cells in the Y-axis. Image A shows the input raw image, images B, C, D show the segmentation results for different values of h. Image B shows over-segmentation, image D shows under-segmentation. The correlation between the minimum variance in the areas of the cells and appropriate segmentation is clearly demonstrated in image C. or faulty tracking of cells. On the other hand, watershed segmentation algorithm finds the geometry and other observed properties of the plant cells more accurately than the level-set, thus producing significantly better correspondence results in tracking (see Fig.5).
Tracking Stem Cells
Graphical abstraction is created on a collection of cells in an image. In this process every cell is characterized by a vertex in the graph and neighboring vertices are connected by an edge. The structure of these graphs automatically contains the relative distance between two neighboring

Figure 5 : Raw image in the first column is segmented using both watershed and level-set segmentations. Second column displays watershed segmentation, while the third column is the result of level set segmentation. Qualitatively, watershed far surpasses level-set segmentation as demonstrated in this figure. cells (the edge length) and the edge orientation. The topology and the geometry of the local graphs should not change if there is no cell division or the images are not noisy. Taking into account above conditions, the correspondences between cells are identified by matching the local graphs (Fig. 6 (A)). To find a correspondence between cells ci and cj across different time instants t and t+1, the

Figure 6 : Image A: The matched local graphs G1 at t and G2 at t + 1 time points, the correspondence of the seed cell pair (1,1’), as well as the correspondences of the neighboring cells, such as (2,2’) and (4,4’). Image B: A diagram to describe the relative position of the mother cell and the daughter cells. distance measure between two local graphs is defined as follows :

where  is a neighboring cell of
is a neighboring cell of  , and
, and  is a neighboring cell of
is a neighboring cell of  , N(c) is the neighboring cell set (set of cells that are within a certain distance around cell c),
, N(c) is the neighboring cell set (set of cells that are within a certain distance around cell c),  ;
; and
and ;
; are the edge lengths,
are the edge lengths,  ;
; and
and  ;
; are the orientation angles in radians of the edges measured relative to a horizontal axis,
are the orientation angles in radians of the edges measured relative to a horizontal axis, and
and  are the cell position vectors, and is the average distance between two neighboring cells. If two local graphs match, which is when the distance measure is small, we can say that the central cells ci and cj are a corresponding cell pair.Overall, tracking of stem cells from any two consecutive cell image stacks using local graph matching
are the cell position vectors, and is the average distance between two neighboring cells. If two local graphs match, which is when the distance measure is small, we can say that the central cells ci and cj are a corresponding cell pair.Overall, tracking of stem cells from any two consecutive cell image stacks using local graph matching