Introduction
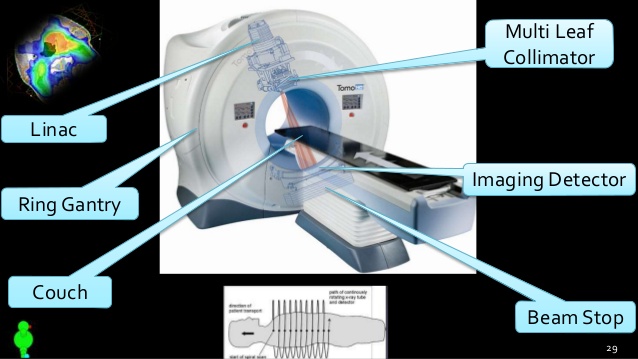
Radiation therapy utilizes ionizing radiation with the goal of curing or palliating disease and minimizing damage to healthy tissue. Localization using Image Guided Radiation Therapy (IGRT) is performed in order to ensure tumor location and minimize toxicity of normal tissue due to inaccurate irradiation.IGRT can consist of both radiation sources or non-radiation sources to track the tumor position. For instance, prior to treating lung tumor using SBRT, a CBCT will be taken to localized the tumor. When examining tumors that are in areas affected heavily by motion,such as lung and abdominal cancers, motion management techniques
are employed including:
- Tracking
- Respiratory gated radiotherapy
Motion management uses real-time imaging and surrogate monitoring to assure that tumor motion is within the radiation eld during high-dose and high-precision radiotherapy.Real-time imaging methods include audio prompting or visual feedback to create relatively reproducible breathing patterns. Another technique utilizes internal organ motion tracking, such as of the diaphragm, to reconstruct respiratory motion. Similarly, breath-hold methods require patients to x their respiratory motion in a specied phase while administering radiation, but this can be troublesome for patients who cannot hold their breath. Other methods include use 4DCT to determine a Sliced Body Volume (SBV) to create a signal similar to an amplitude motion signal In regions where tumor motion is heavily affected by respiration, radiation gating is used to precisely target the tumor during beam-on time. Respiratory gating radiotherapy requires the administration of radiation at particular points in a patients respiratory phase.Respiratory gating radiotherapy utilizes external signals obtained from systems such as the Real-time Position Management TM (RPM) System, internal signals from systems such as the Calypso Tracking, or external radiation such as uoroscopy to determine when the radiation beam should be turned on because the tumor is in the eld.
Neural Networks
Other methods utilize Neural Networks (NN) to create a data base of patterns and possible predictions. NN require a certain learning speed to create such a database. In Yan’s technical study using a NN, surgical clips were required to determine if internal motion can be predicted from external markers. The study uses multiple markers and several data must be developed to produce a complex network for prediction. Additionally, as samples for predictions were increased over 6 seconds there was a noticeable negative effect on the correlation between predicted signal and internal signal and the predicted error between the prediction and internal signal. Similarly, Seregni’s feasibility study uses a NN to track tumors but the nal outcome has some amplitude errors which reach up to 5mm, which is insufcient for high-precision radiosurgery.
Circular Pattern Augmentation
Hong’s methods use angular velocity and circular motion to make predictions. The circular nature of respiratory motion is exhibited by the CIRS phantom and replicating this in respiratory prediction seems obvious. However, Hong only attempts to make predictions for 0.4 s, which would only account for RPM system latencies and would be inecient for radiosurgery
Pattern Fusion
Pattern fusion prediction is novel because it has been considered for cloud storage prediction and update forecasting and will now be applied for tracking and gating prediction. Pattern fusion will take previously collected data from a patient surrogate signal to fuse together creating a prediction. Surrogate motion position is adequate information because demonstrated previously is strong correlation between internal position and external surrogate markers.
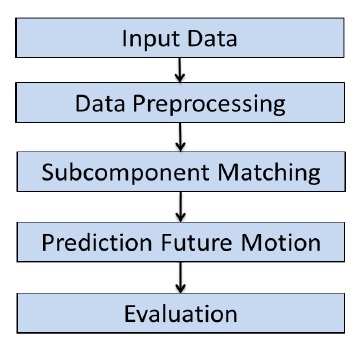
Figure 1: General Prediction Algorithm Workflow.
The prediction algorithm work ow has been designed based on Yang et. al. Pattern Fusion Model for Multi-Step-Ahead CPU Load Prediction. The algorithm work ow is depicted in Figure1 as a owchart. Each step in the algorithm